SKIPPY
A Tool for the Detection of Exonic Variants that Modulate Splicing
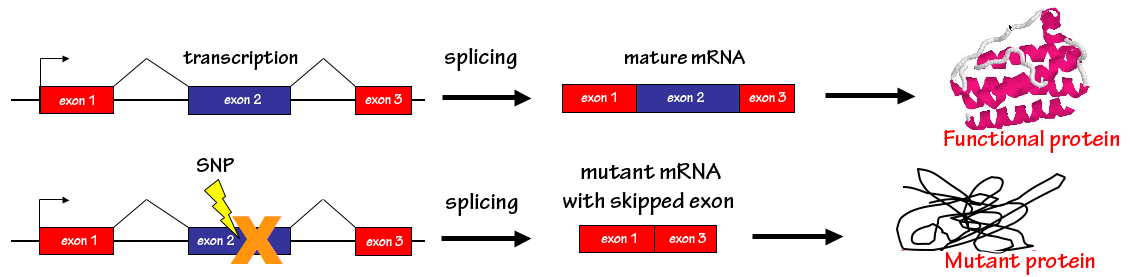
Missense, nonsense and synonymous mutations deep within exons, but outside of the splice junctions (i.e. >3bp internal to the exon) can have devastating effects on gene function by causing exon skipping or activating ectopic splice sites. The confounding location of these mutations in mostly coding sequence, as well as the lack of a clear strategy for their identification, means that their potential effects on splicing are often overlooked.
Skippy is a Web-based tool that allows users to input a set of exonic variants to score them for a number of features (such as changes in splicing regulatory elements) that have been shown to be predictive of known genome variations that cause exon skipping or activation of ectopic splice sites. In this way, variants can be either prioritized for further splicing-based functional analysis or the results can be used as further genomic evidence in cases in which the causative variant is already known.
For more details please read our accompanying paper:
- Woolfe, A., Mullikin, J., and Elnitski, L. 2009. Genomic features defining exonic variants that modulate splicing. Genome Biology. 11:R20 [PubMed]
Exonic variants can be submitted to the program here
Regulatory Constraint (RC) Score Wiggle Track
The RC score is a measure of how likely the observed level of conservation is, in a region of an exon, given the underlying codons. Here, the algorithm is implemented specifically to measure possible additional evolutionary constraint on the coding sequence caused by the presence of important splicing regulatory elements. More details on how this score is obtained can be found here. The Wiggle track for scores for all positions in internal coding exons in the UCSC Genome Browser (assembly hg18) can be downloaded here (~43.5Mb). If you experience problems downloading the file, please right-click on the link and select 'Download linked file as...'. Once the file is downloaded, untar and unzip the files, and then wiggle tracks for each chromosome can be uploaded as a 'custom track' here.