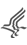Exonic Environment and Intronic ESS Density
Summary
- While known SAVs in general dont have significantly lower splice site strengths, other studies show that weak splice sites can make an exon dependant on internal splicing regulatory elements, and are therefore more vulnerable to SAVs
- Exons containing SAVs tend to have lower densities of ESEs and higher densities of ESSs, possibly reflective of weak exon definition
- Introns flanking exons containing SAVs tend to have higher ESS densities, again suggestive of greater dependance on splicing regulatory elements within the exon, for exon definition
Exonic Environment
Splice Site Strength
Although are comparative analysis of SAVs did not find that exons containing SAVs have significantly lower splice site strengths than those of the same size containing hSNPs or even exons in general, it is known from a large body of work that splice site strengths do influence exon definition and cause exons to have greater dependance on, or be influenced by nearby splicing regulatory elements.
In Skippy, we use the MaxEnt program to score splice site strengths. Although no thresholds are set to designate splice sites as 'strong' or 'weak' in the MaxEnt program, we set a threshold score of 5 and below to designate 'weak' sites for 5' and 3' splice sites. This score represents 1.8 standard deviations lower than the mean scores of all constitutively spliced exons genome-wide. We shade scores at or below the threshold (i.e. weak splice sites) in red while scores above the threshold are shaded blue.
Exonic ESE/ESS Density
Another major feature thought to distinguish exon sequences from their surrounding intronic environment is the prevalence of higher densities of ESEs and low or absent densities of ESSs, (while introns and pseudoexons may be defined in the exact opposite terms).
In a comparison of exons containing SAVs against a random boot-strap sampling approach of exons (of similar size distributions) containing hSNPs and of constitutive exons genome wide, we found that SAV-exons have significantly lower ESE densities and significantly higher ESS densities than these other category of exons. The ESE/ESS densities of SAV exons represent a shift away from strong exon definition and towards ESR densities observed within introns. This weakened exon definition, may explain the vulnerability of these exons to variants that further modulate the ESE/ESS density which leads to exon skipping.
As most of the important splicing signals are thought to be close to the splice junctions we meaure ESR densities in the first and last 50bp of the exons. For exons that are less than 100bp, densities are measured across the entire length of the exon. To allow users to compare their exon densities with those of hSNP exons of a similar size, Skippy shades exonic ESE densities red that are less than the mean hSNP exonic ESE density and blue densities that are higher than the mean hSNP exonic ESE density.
For ESS densities, values are shaded red if they are higher than the mean hSNP exonic ESS density and blue if they are less.
Intronic Environment
Intronic ESS Density
We found higher densities of ESSs in the intronic sequence directly flanking SAV exons. It is not clear that ESSs act as the same functional elements when present in an intronic context, as they may act as both intronic splicing enhancers (ISEs) and silencers (ISSs) when placed in an intronic context. If ESSs in an intronic context do act mainly as ISSs, they may have a negative influence on the nearby splice sites in a way that makes the exon increasingly reliant on splicing enhancers within the exon. We measure ESS densities in the first 100bp upstream and values are 100bp downstream and shaded red if they are higher than the mean hSNP intronic density and blue if they are less.